Paris, France + Partial Remote
https://avatarmedical.ai/

AVATAR MEDICAL is an exciting new MedTech start-up that radically enhances how surgeons
prepare their surgeries.
Based on technology developed at the Institut Pasteur and Institut Curie, AVATAR MEDICAL
uses advanced image processing algorithms coupled with extended reality (XR) to transform
medical images into lifelike patient avatars. This vehicle grants surgeons, patients, and students
an intuitive and powerful means of understanding the intricacies of medical imaging data.
AVATAR MEDICAL is a laureate of the prestigious BPI i-Lab, BPI i-Nov, and EIC Accelerator
competitions for innovative French startups and was the winner of the Laval Virtual 2022 Grand
Prix.
We are offering an exciting Computer Vision internship opportunity within our technology
Innovation Team. The successful candidate will work on a challenging project consisting in the
development of a mixed reality solution to train surgeons with specific surgical gestures. This is
a truly multidisciplinary project that combines medical imaging, image processing and
high-fidelity computer vision techniques for a distinct clinical training purpose.
Missions
● Work with surgeon partners to assess surgical training needs
● Assess different computer vision approaches for surgical use case
● Develop mixed reality surgical training prototype(s)
● Document the testing and development phases of said prototype
Required Skills
● Experience with computer vision approaches (e.g., OpenCV, SLAM, etc)
● Proficiency in C, C++ and Python programming languages
● Strong understanding of mathematics and linear algebra
● Confirmed problem-solving skills and ability to work well in a team
● Fluency in English (French language proficiency is not mandatory)
Nice to Have (not required, but a plus):
● Experience with Unity Engine and/or Unreal Engine
● Familiarity with AI object detection architectures (e.g., YOLO)
● Understanding of object-oriented design and SOLID principles
Why Join Avatar Medical?
● 30+ team members representing 7 nationalities, with offices in Paris, Saint-Malo, and
California
● Collaborate with talented and passionate professionals from around the world
● Join a fast-growing company at the intersection of technology and healthcare
● A competitive internship compensation package above the legal minimum
● International work experience
● Meal vouchers worth €10 per day, with €6 covered by the company
● Flexible hybrid work policy
Contract
Duration: Internship 6 month, from January 2025
Parisante Campus 10 rue d’Oradour Sur Glane 75015 Paris
Contact
If you are interested and qualified for this opportunity, please contact jobs@avatarmedical.ai
with the following:
● CV
● Motivation Letter
Drosophila larvae have emerged as an ideal platform for simultaneously probing behaviour and the underlying neuronal computation1–5. Modern genetic tools allow efficient activation or silencing of individual and small groups of neurons. Combining these techniques with standardised stimuli over thousands of individuals makes it possible to relate neurons to behaviour causally However, extracting these relationships from massive and noisy recordings requires the development of new statistically robust approaches.
Recently, in experimental settings to probe defensive actions6 or understand the implementation of neuromodulation in small neural neworks7, larva populations exhibited significant deviations from usual behavioural features. These deviations lead to either the redefinition of features usually associated with these behaviours or to challenges in detecting them.
We aim to develop a Bayesian Program Synthesis8 (BPS) methodology for producing synthetic data that mirrors key characteristics found in experimental recordings. This method will entail an inference phase to identify large-scale parameters characteristic of larva populations. It will incorporate various generative models to either replicate the behavioural traits associated with particular genotypes or create bespoke behavioural features. Additionally, we plan to use a collection of small generative programs to create larva data under specific action conditions. The efficacy of this approach will be demonstrated by applying our behaviour analysis pipeline to the synthetic data and assessing its ability to facilitate transfer learning with annotated behavioral datasets.
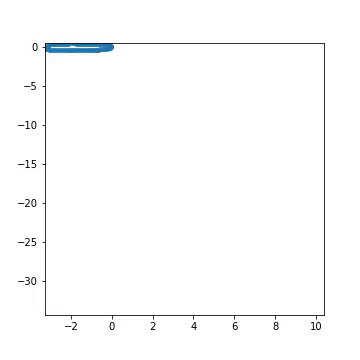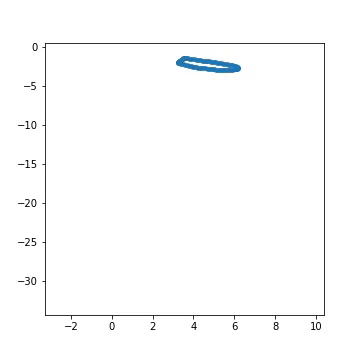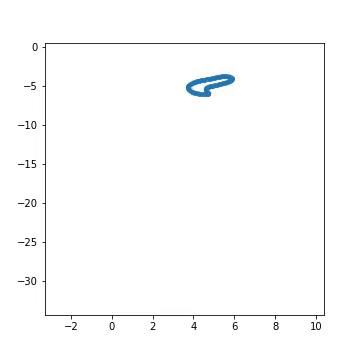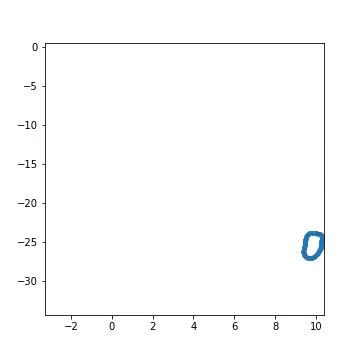
Fig 1) Example of artificial generated from a pre-defined larva genotype
References
1. Masson, J.-B. et al. Identifying neural substrates of competitive interactions and sequence transitions during mechanosensory responses in Drosophila. PLOS Genet. 16, e1008589 (2020).
2. Jovanic, T. et al. Competitive Disinhibition Mediates Behavioral Choice and Sequences in Drosophila. Cell 167, 858-870.e19 (2016).
3. Croteau-Chonka, E. C. et al. High-throughput automated methods for classical and operant conditioning of Drosophila larvae. eLife 11, e70015 (2022).
4. Vogelstein, J. T. et al. Discovery of Brainwide Neural-Behavioral Maps via Multiscale Unsupervised Structure Learning. Science 344, 386–392 (2014).
5. Winding, M. et al. The connectome of an insect brain. Science 379, eadd9330 (2023).
6. Lehman, M. et al. Neural circuits underlying context-dependent competition between defensive actions in Drosophila larva. 2023.12.24.573276 Preprint at https://doi.org/10.1101/2023.12.24.573276 (2023).
7. Tredern, E. de et al. Feeding-state dependent modulation of reciprocally interconnected inhibitory neurons biases sensorimotor decisions in Drosophila. 2023.12.26.573306 Preprint at https://doi.org/10.1101/2023.12.26.573306 (2023).
8. Lake, B. M., Salakhutdinov, R. & Tenenbaum, J. B. Human-level concept learning through probabilistic program induction. Science 350, 1332–1338 (2015).
Scientific or technical background required for work program
The successful intern should have one of the following backgrounds:
- Statistical Physics, Applied Mathematics,
- Statistics & Bayesian Inference
Some fluency in Python is expected.
Behavior and decision-making are determined by physical processes taking place in the complex environment of the brain. Experimental techniques have reached the point where it is now possible to map the complete wiring diagram (the anatomical connectome) of the brain of simple model organisms at the level of single synapses [1,2,3], and to control the activity of individual neurons in live animals and observe the resulting behavior [4]. Together this offers the occasion to reverse engineer the physical basis of behavior [4].
This project aims to employ computational, statistical, and machine learning methods to characterize the neural circuitry of the neuronal networks of small animals and investigate the influence of physical constraints on their topology. The project involves two subparts, which may be investigated individually or collectively as time allows and depending on the interests of the student:
- In search of biological neural network distances. Comparing networks is an open technical challenge as the multiplicity of existing tools does not bring a definitive answer [5]. For instance, comparing networks of different sizes is a non-trivial problem [5]. Network patterns — motifs [6]— are attractive structural features since their definition is a priori independent of the network size. We recently developed an inferential method that extracts statistically significant microcircuits from connectomes. This part of the project is focused on the comparison of circuit motifs as a proxy for network distances. The student would characterize and compare inferred sets of network motifs by exploring how dynamical models [7] are affected by motifs’ topological properties, e.g. their symmetry. Potential extensions of the subject would involve investigations of how a distribution of microcircuits affect the mesoscopic, or modular, structure of a network [8].
- Generative modeling of biologically-plausible brain networks from learnt latent embeddings. This axis aims to develop new generative network models based on embedding the nodes in a metric space where distances determine link probabilities based on a kernel that is learnt from data. This will involve generalizing latent space graph models [9] by learning the distance kernels defining the connection probabilities between neurons from data and possibly extending them to non-euclidean embedding spaces. In a second time, we aim to link these embeddings to higher-order circuit motifs and to the real 3D space embedding of neurons and other biophysical constraints acting on the neural network.
The internship may lead to a PhD scholarship in the lab to continue the project.
References
- [1] Winding et al., “The connectome of an insect brain.” Science 379: eadd9330 (2023).
- [2] Witvliet et al., “Connectomes across development reveal principles of brain maturation.” Nature 596: 257–261 (2021).
- [3] Dorkenwald et al., “Neuronal wiring diagram of an adult brain.” biorxiv https://doi.org/10.1101/2023.06.27.546656 (2023).
- [4] Jovanic et al., “Competitive Disinhibition Mediates Behavioral Choice and Sequences in Drosophila.” Cell 167: 858–870 (2016).
- [5] Hartle, H., Klein, B., McCabe, S., Daniels, A., St-Onge, G., Murphy, C., & Hébert-Dufresne, L. (2020). Network comparison and the within-ensemble graph distance. Proceedings of the Royal Society A, 476(2243), 20190744.
- [6] Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D., & Alon, U. (2002). Network motifs: simple building blocks of complex networks. Science, 298(5594), 824-827.
- [7] Riascos, A. P. (2023). Dissimilarity between synchronization processes on networks. arXiv preprint arXiv:2309.13163.
- [8] Yamamoto, H., Spitzner, F. P., Takemuro, T., Buendía, V., Murota, H., Morante, C., … & Soriano, J. (2023). Modular architecture facilitates noise-driven control of synchrony in neuronal networks. Science advances, 9(34), eade1755.
- [9] Hoff et al., “Latent Space Approaches to Social Network Analysis.” JASA 1-00: 286–295 (2002).
Scientific or technical background required for work program
The successful intern should have training in one of the following fields: statistical or condensed matter physics, applied mathematics, or statistics.
Some fluency in Python and numerical simulations is expected.
